The videos and images below are Collected from experiments in the Cancer Engineering Laboratory
Note: Some of the images below are a taken from movies created from time-lapse experiments – links to the movies are provided at the bottom of the text descriptions.
 |
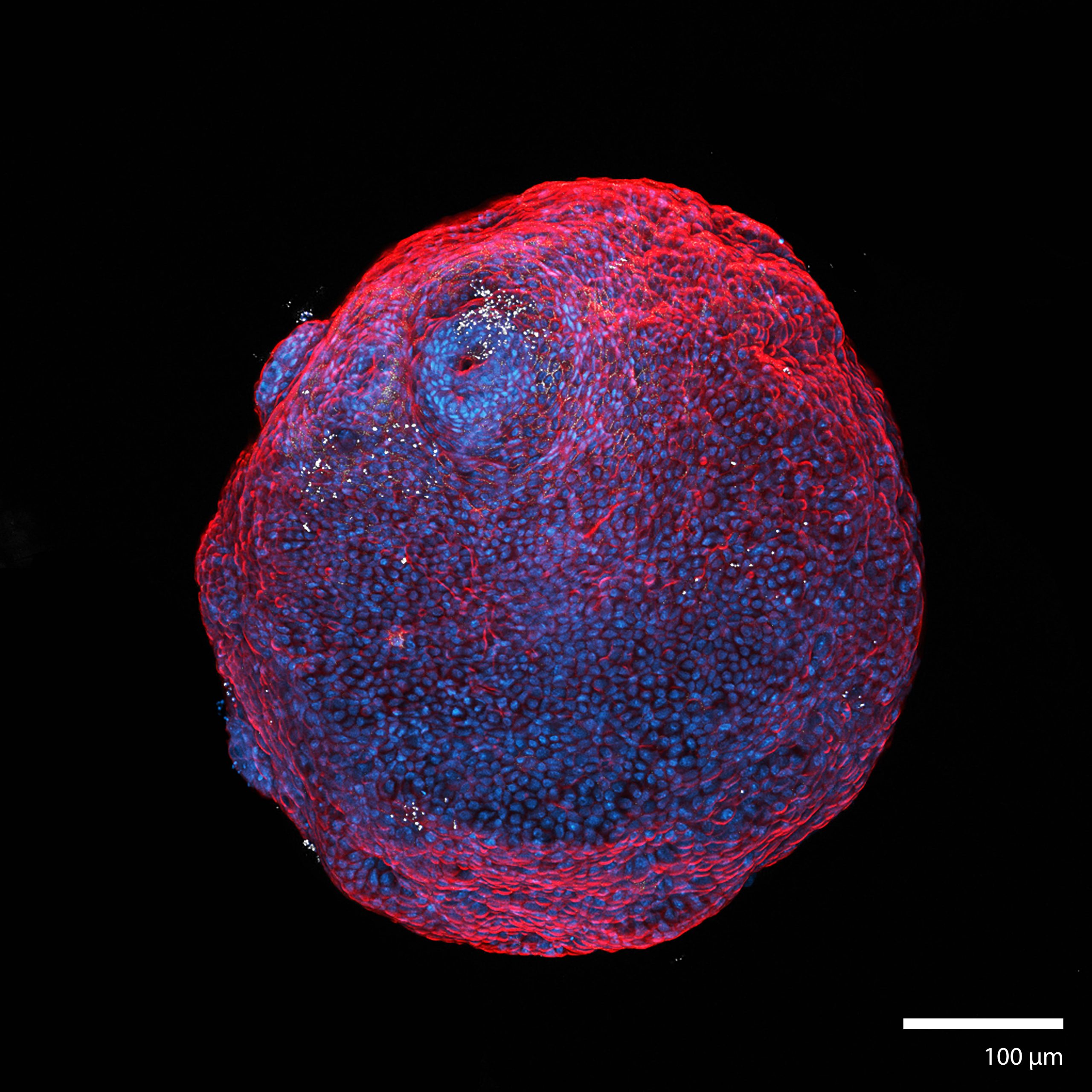
3D Human Heart MicrotissuesThis microtissue was formed from a mechanically disaggregated sample taken during the implantation of a lower ventricular assist device. At the time of imaging the sample was 7 days old, and had been under continuous perfusion in our Darcy plates (see below) at 90% background oxygen partial pressure. The samples were used during the COVID-19 Pandemic to study SARS-CoV-2 infection and interactions with cardiac tissues. Viability was confirmed out to 45 days, although sample integrity was greatly diminished after 28 days. These heart microtissue samples were roughly 400 um in diameter (largest dimension), and some began coordinated contractions after 3 days in culture. This heart microtissue sample was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 7 days and stained for actin (TRITC: red), the nuclei (DAPI: blue), and for Troponin (FITC: green). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
3D Colorectal OrganoidsThis colorectal organoid was formed from a mechanically disaggregated sample of the large intestine. The microtissue fragment was initially faceted as a result of the mechanical disaggregation, but became smooth and spherical during a maturation process under perfusion in the 3D Darcy Plates. At the time of imaging the sample was 14 days old, and had been under continuous perfusion in our Darcy plates at 90% background oxygen partial pressure. This sample was used during an exploratory study of liquid culture medium tuning and optimization. Viability was confirmed out to 21 days, at which time the experiments were stopped; the samples were still functional and viable. The intestinal crypts formed on these organoids, which can be seen as the circular protuberances on the periphery. These colorectal organoid samples were roughly 200-400 um in diameter. The colorectal organoids produced copious amounts of mucin, and some showed prolonged coordinated contractions over weeks in 3D culture. This colorectal organoid was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 14 days and stained for actin (TRITC: red), the nuclei (DAPI: blue), and E-cadherin (Cy5: white). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
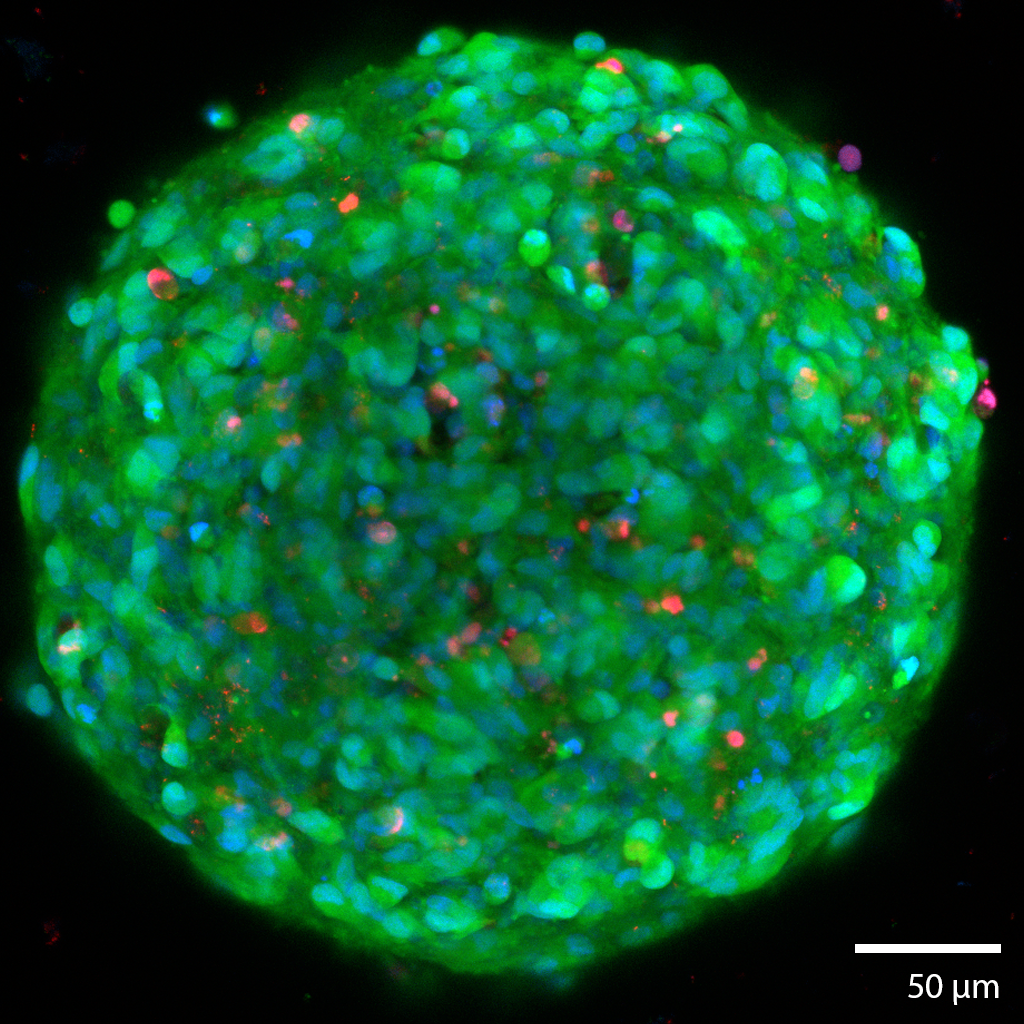 |
3D Glioma TumoroidsThis glioma cancerous “tumoroid” was formed from a mechanically disaggregated sample taken during a biopsy of the glioma. The microtissue fragment was initially faceted as a result of the mechanical disaggregation with large amounts of necrotic tissue. These spherical tumoroids grew out of the original microtissue fragments over 3 days under perfusion in the 3D Darcy Plates. At the time of imaging this sample was 14 days old, and had been under continuous perfusion with Neurocult liquid media with supplements and 5% fetal bovine serum in our Darcy plates at 90% background oxygen partial pressure. This sample was used as part of our in vitro experiments in immuno-oncology. The tumoroids all grew to approximately 250 um in diameter, and all displayed similar sphericity and intact morphology. This gioma tumoroid is stained with a viability stain Calcein-AM (FITC: green), a nuclear stain Hoechst- 33342 (DAPI: blue), and a dead stain BOBO-3 Iodide (TRITC: red). Small necrotic fragments were found throughout the 3D medium from the beginning, and is characteristic of gliomas. Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
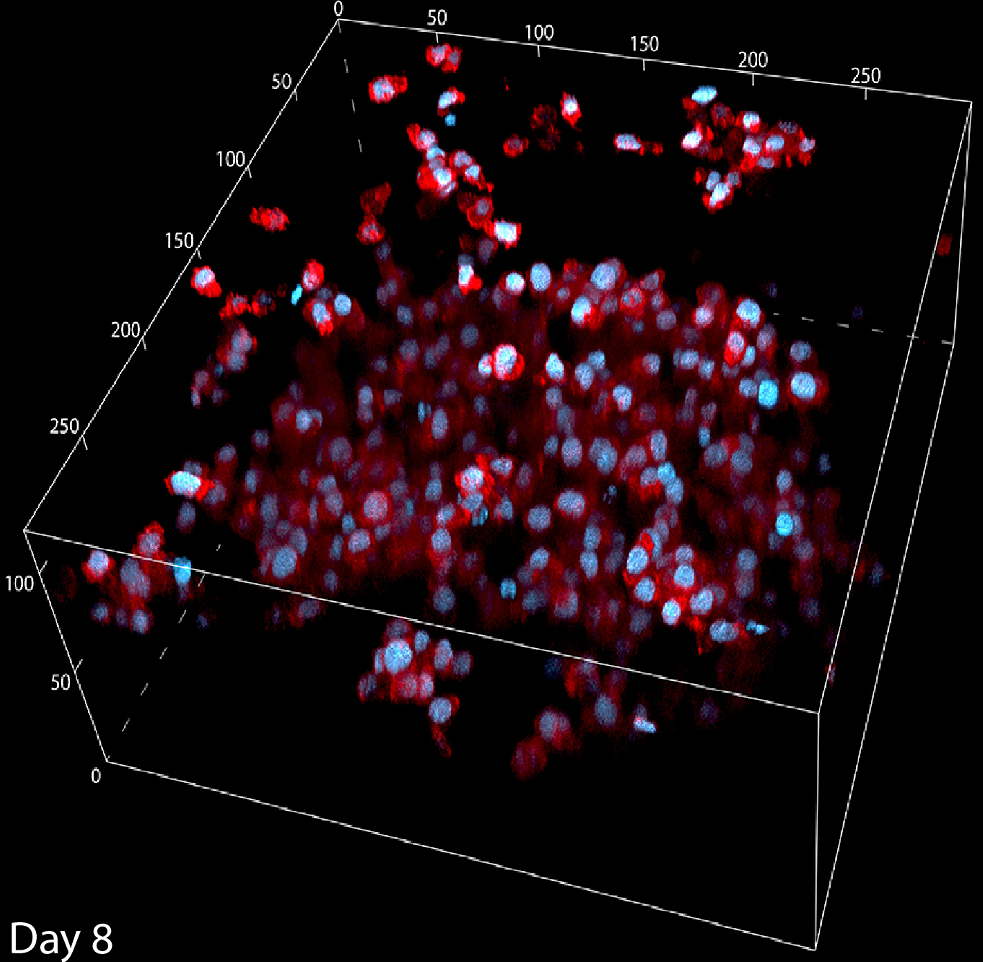
activated human immune cells in 3DThis collection of immune cells have been expanding over 2 weeks under perfusion in the 3D Darcy Plates. At the time of imaging this aggregated collection of immune cells had been in culture for 8 days under continuous perfusion with RPMI 1640 with 10% human serum and 1% Glutamax with 60U/ml IL2. Background oxygen partial pressures were approximately 20%. These cells are used as part of our in vitro experiments in immuno-oncology, and are taken from collections of donor blood. Activation occurs in the Liquid Like Solid (LLS) 3D Culture medium using Dynabeads coated with aCD3 and aCD28 antibodies. The beads are stationary within the LLS medium, and the immune cells move through interstitial space in the LLS, interact with the beads, proliferate, and expand in regional aggregates. This immune cell aggregate was fixed and stained in situ in the LLS. F-Actin is stained with AlexaFluor 555 (TRITC:red), and the nuclei were stained with NucBlue (DAPI: blue) Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
3D In vitro Immuno oncology experiment
|
|
3D In vitro Immuno oncology experiment
|
|
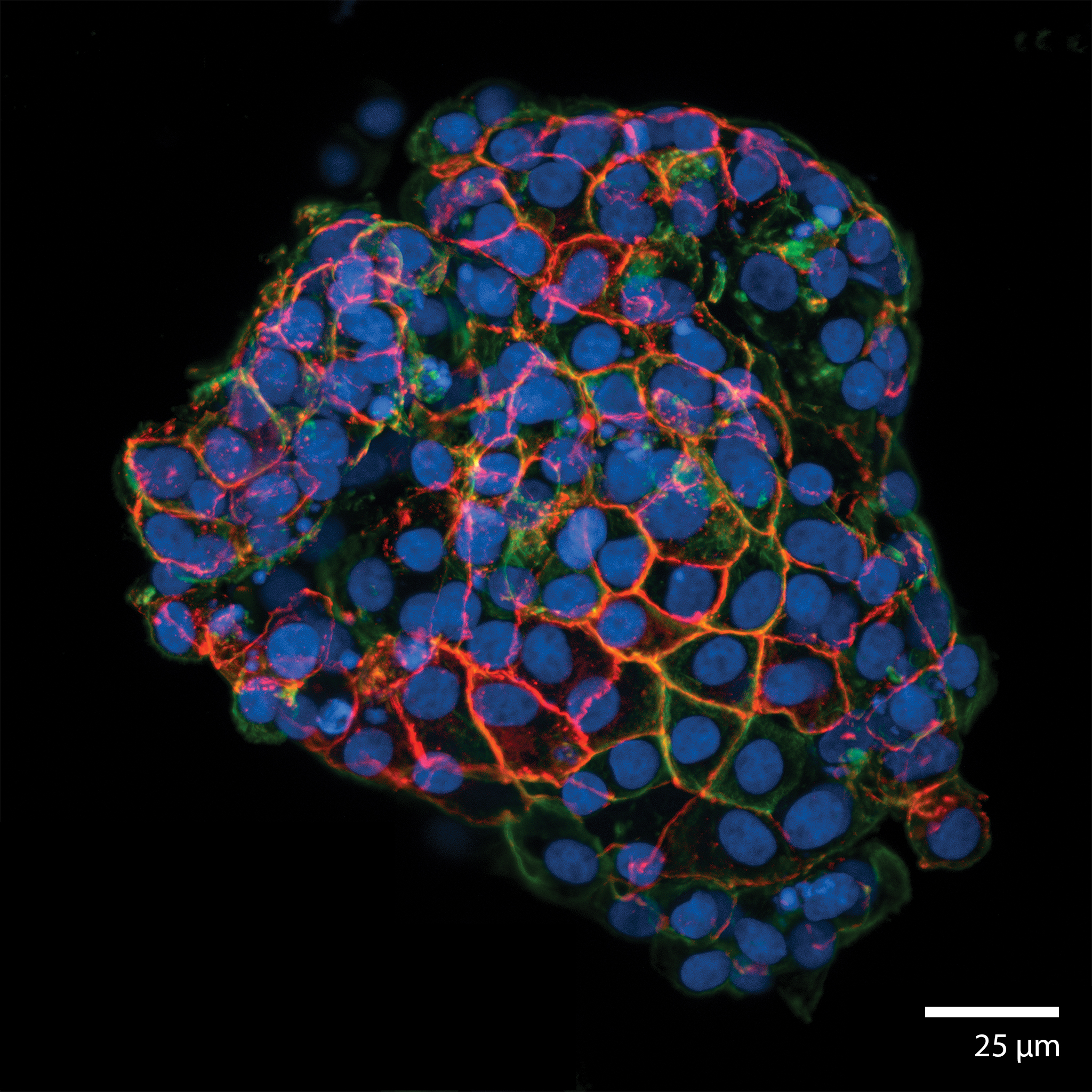 |
intestinal epithelial cell (iec-6) organoidsThis image is taken from an experiment 3D printing (see: “Writing in the Granular Gel Medium) IEC-6 cells (cell-line) and matured over 7 days under continuous perfusion in our Darcy plates at 20% background oxygen partial pressure. This sample was used during an exploratory study of organoid biofabrication. These intestinal organoids grew to roughly 200-400 um in diameter. These organoids were extremely durable, and showed viability out to over 60 days, even in the absence of perfusion. The cells showed strong motility along the surface of the organoid, which could be followed via in situ scanning confocal microscopy in the LLS medium. This organoid was stained with for tight junction protein-1 (ZO-1) (TRITC:red), actin (FITC:green), and the nuclei were stained with NucBlue (DAPI:blue) Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
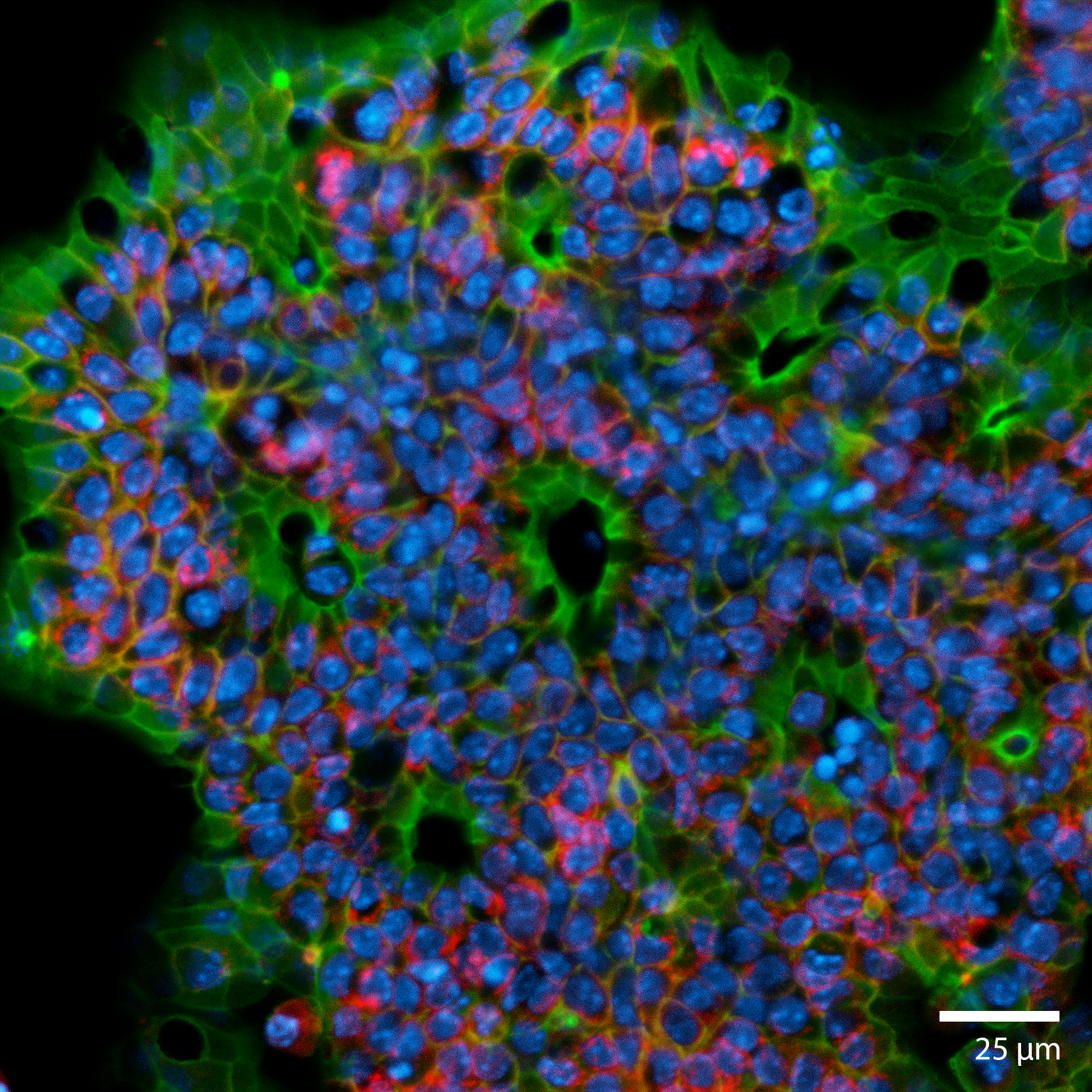
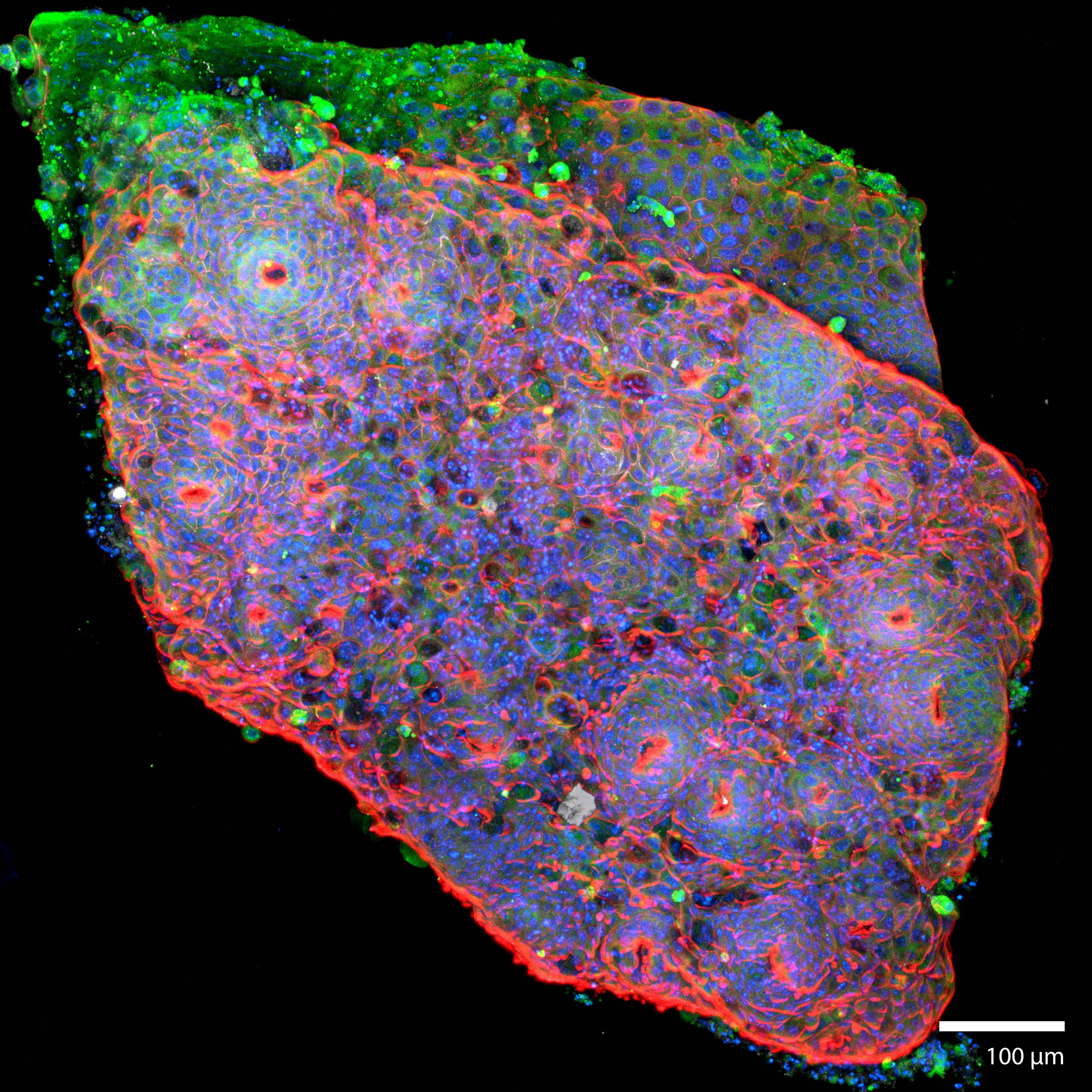
3D Colorectal OrganoidsThis image is taken from a colorectal organoid that was formed from a mechanically disaggregated sample of the large intestine. The microtissue fragment was initially faceted as a result of the mechanical disaggregation, but became smooth and spherical during a maturation process under perfusion in the 3D Darcy Plates. At the time of imaging the sample was 7 days old, and had been under continuous perfusion in our Darcy plates at 90% background oxygen partial pressure. This image is a confocal scanned cross-section of the surface in an effort to visualize the intestinal crypts. This colorectal organoid was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 7 days and stained for F-actin using Phalloidin (FITC: red), the nuclei Hoechst 33342 (DAPI: blue), and E-cadherin (Cy5: shown red). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
 |
3D Colorectal microtissue explants and 3D OrganoidsThis colorectal organoid was formed from a mechanically disaggregated sample of the large intestine. The microtissue fragment retained its initially faceted morphology from mechanical disaggregation during a maturation process under perfusion in the 3D Darcy Plates. At the time of imaging the sample was 14 days old, and had been under continuous perfusion in our Darcy plates at 90% background oxygen partial pressure. This sample was used during an exploratory study of liquid culture medium tuning and optimization. Viability was confirmed out to 21 days, at which time the experiments were stopped; the samples were still functional and viable. The intestinal crypts were retained on these organoids, which can be seen as the circular protuberances on the face of the microtissue explants. These colorectal organoid samples were roughly 300-500 um in largest dimension. These colorectal organoids and microtissue explants produced copious amounts of mucin, and some showed prolonged coordinated contractions over weeks in 3D culture. This colorectal organoid was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 14 days and stained for for tight junction protein-1 (ZO-1) (FITC:green), actin Phalloidin (TRITC:red), the nuclei were stained with the nuclei Hoechst 33342 (DAPI: blue), and E-cadherin (Cy5: white). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
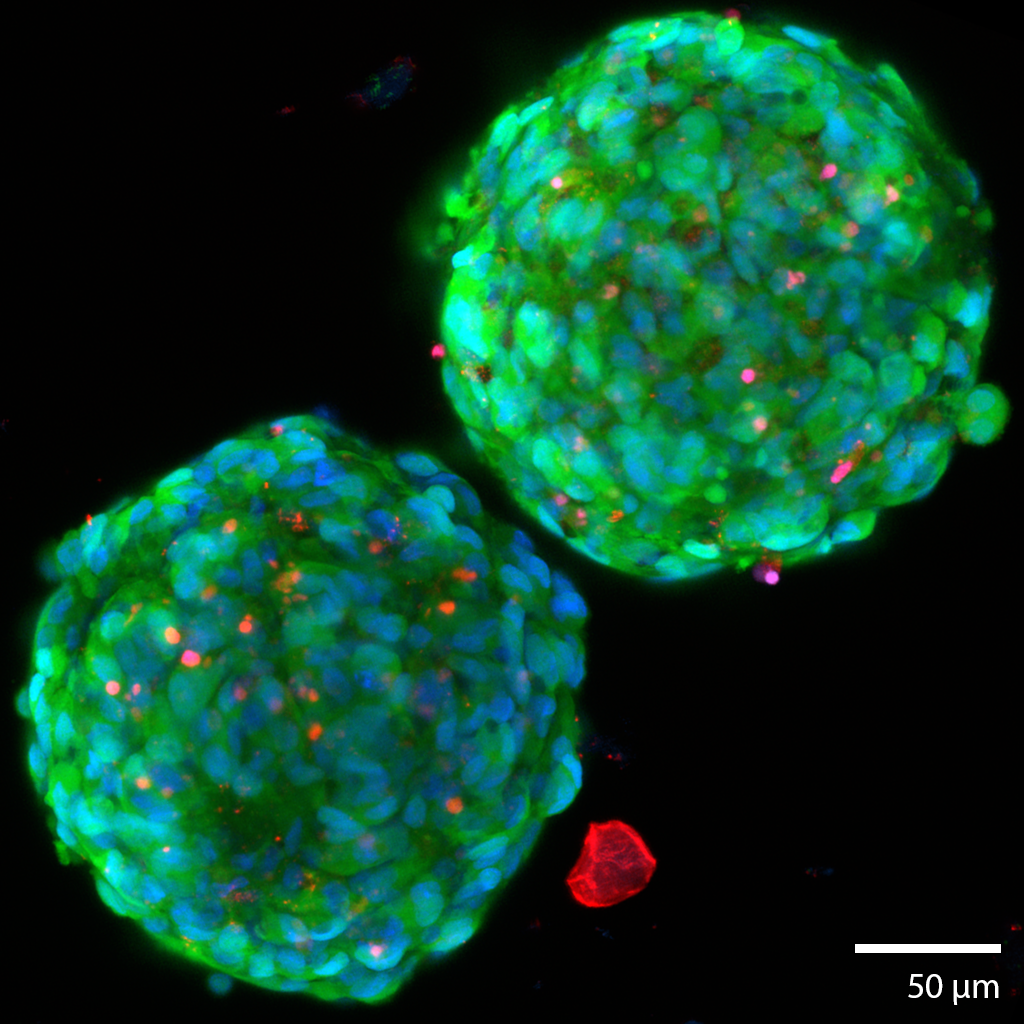
3D Glioma TumoroidsThis pair of glioma cancerous “tumoroids” were formed from a mechanically disaggregated sample taken during a biopsy of the glioma. The microtissue fragment was initially faceted as a result of the mechanical disaggregation with large amounts of necrotic tissue. These two spherical tumoroids grew out of the original microtissue fragments over 21 days under perfusion in the 3D Darcy Plates. Continuous perfusion with Neurocult liquid media with supplements and 5% fetal bovine serum in our Darcy plates at 90% background oxygen partial pressure. These sample was used as part of our in vitro experiments in immuno-oncology. The tumoroids all grew to approximately 250 um in diameter, and all displayed similar sphericity and intact morphology. This gioma tumoroid is stained with a viability stain Calcein-AM (FITC: green), a nuclear stain Hoechst- 33342 (DAPI: blue), and a dead stain BOBO-3 Iodide (TRITC: red). Note the necrotic fragment in the vicinity of the tumoroids, similar fragments were found throughout the 3D medium from the beginning, and are characteristic of gliomas. Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
3D In vitro Immuno oncology experiment
|
|
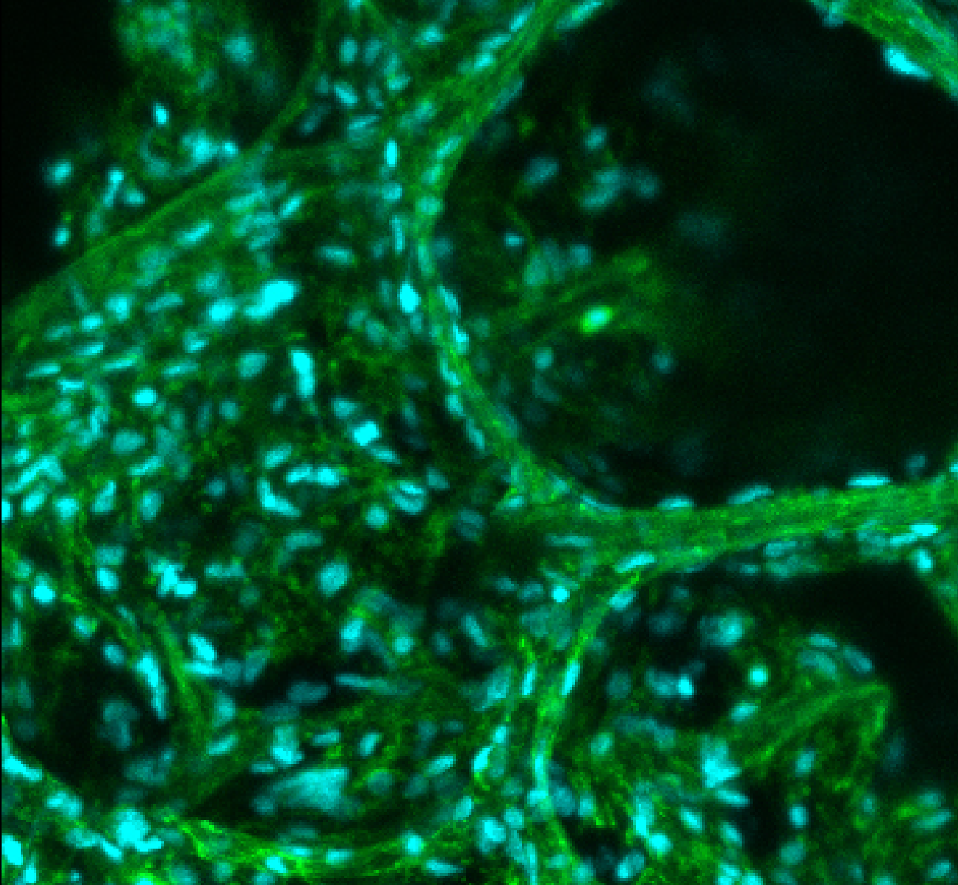
3D Human Lung Microtissue Model for COVID-19 and sars-cov-2 experimentsThis microtissue is from a mechanically disaggregated sample taken from a non-cancerous region of lung after a lobectomy. The aveolar space and structure were maintained as well as cellular heterogeneity and resident immune cells. At the time of imaging the sample was 7 days old, and had been under continuous perfusion in our Darcy plates (see below) at 90% background oxygen partial pressure. The samples were used during the COVID-19 Pandemic to study SARS-CoV-2 infection and interactions with pulmonary tissues. Viability was confirmed out to 14 days, although most infection and drug study experiments took only 72 hours. These lung microtissue samples were roughly 400 um in largest dimension. This lung microtissue sample was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 7 days and stained for F-actin Phalloidin (FITC: green) and the nuclei were stained with Hoechst 33342 (DAPI: blue). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
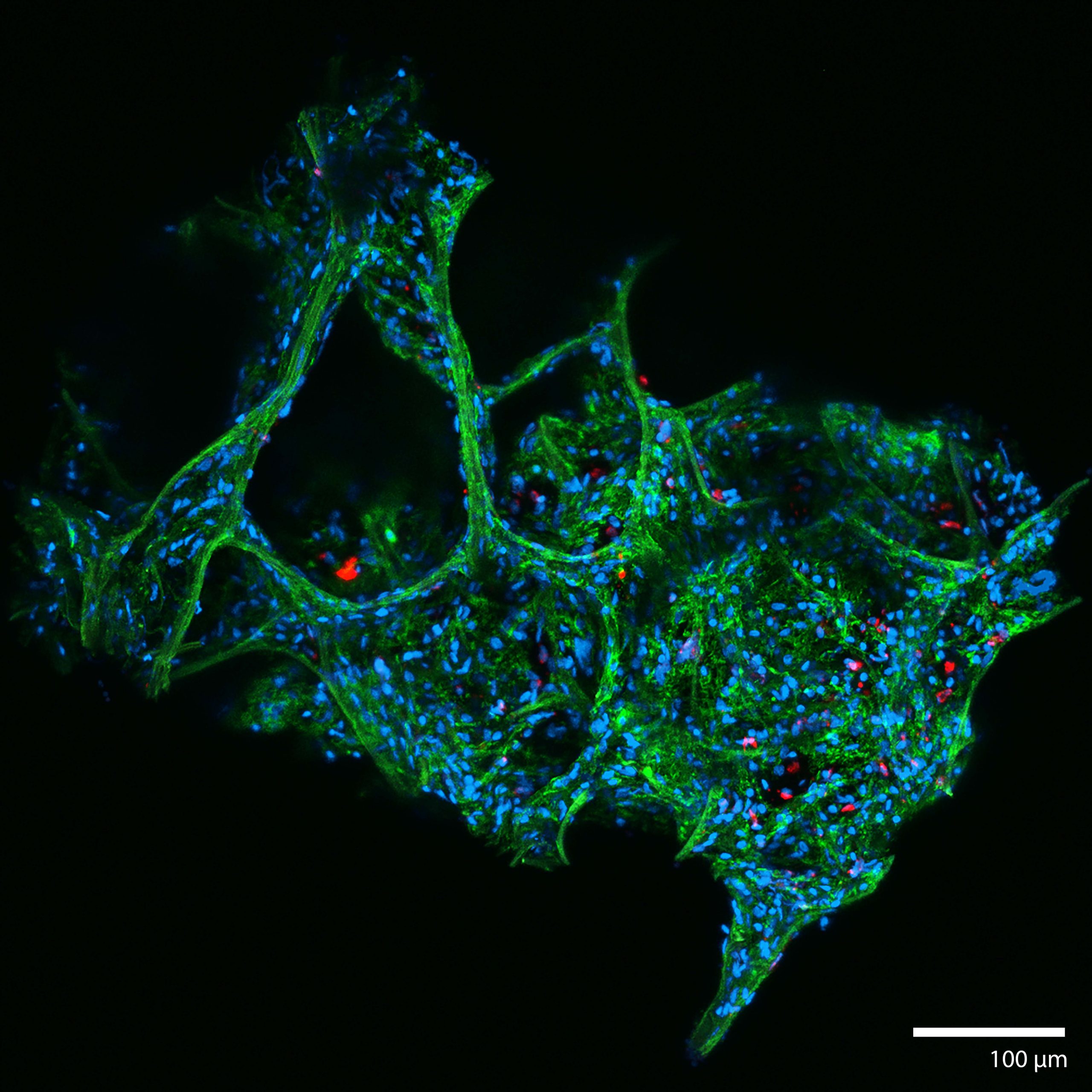 |
3D Human Lung Microtissue Model for COVID-19 and Coronavirus experimentsThis microtissue is from a mechanically disaggregated sample taken from a non-cancerous region of lung after a lobectomy. The aveolar space and structure were maintained as well as cellular heterogeneity and resident immune cells. At the time of imaging the sample was 7 days old, and had been under continuous perfusion in our Darcy plates (see below) at 90% background oxygen partial pressure. The samples were used during the COVID-19 Pandemic to study SARS-CoV-2 infection and interactions with pulmonary tissues. Viability was confirmed out to 14 days, although most infection and drug study experiments took only 72 hours. These lung microtissue samples were roughly 400 um in largest dimension. The infection here, shown in TRITIC:red is from a human coronavirus OC43 (HCoV-OC43), and was performed in our BSL2 facility. This lung microtissue sample was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 7 days and stained for F-actin Phalloidin (FITC: green) and the nuclei were stained with Hoechst 33342 (DAPI: blue), and HCoV-OC43 was stained with TRITC:red. Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
3D Human Lung Microtissue Model for COVID-19 and sars-cov-2 experimentsThis microtissue is from a mechanically disaggregated sample taken from a non-cancerous region of lung after a lobectomy. The aveolar space and structure were maintained as well as cellular heterogeneity and resident immune cells. At the time of imaging the sample was 7 days old, and had been under continuous perfusion in our Darcy plates (see below) at 90% background oxygen partial pressure. The samples were used during the COVID-19 Pandemic to study SARS-CoV-2 infection and interactions with pulmonary tissues. Viability was confirmed out to 14 days, although most infection and drug study experiments took only 72 hours. These lung microtissue samples were roughly 400 um in largest dimension. The infection here is with SARS-CoV-2, which was performed in a BSL3 facility. This lung microtissue sample was fixed in place and stained for F-actin Phalloidin (TITC: red). the nuclei were stained with Hoechst 33342 (DAPI: blue), and SARS-CoV-2 was stained with FITC:green. Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
 |
DARCY plates3D perfusion plates for long term culture of organoids, microtissue explants, and in vitro immuno ocology experiments.This image is from a time lapse movie showing perfusion of the liquid media through the Liquid Like Solid (LLS) 3D culture medium. The Darcy Plates use a passive negative pressure (sub ambient pressure) device to draw the liquid fluid through the LLS and around the tissue and cells in the LLS and into 4 isolated collection wells. The plate has 24 individual experiments (24-wells) group in sets of 6 (4 quadrants) and allow for continuous and steady perfusion. The effluent can be collected through a collection port without opening the Darcy place. The gives passage free continuous culture of 3D tissues, organoids, and experiments with immune cells and drug screening. The Darcy plates enable our immuno-oncology experiments and long duration 3D culture. Although the Darcy plates are smaller than traditional 96-well plates, they work within standard cell culture infrastructure. movie: Darcy Plates 3D Perfusion |
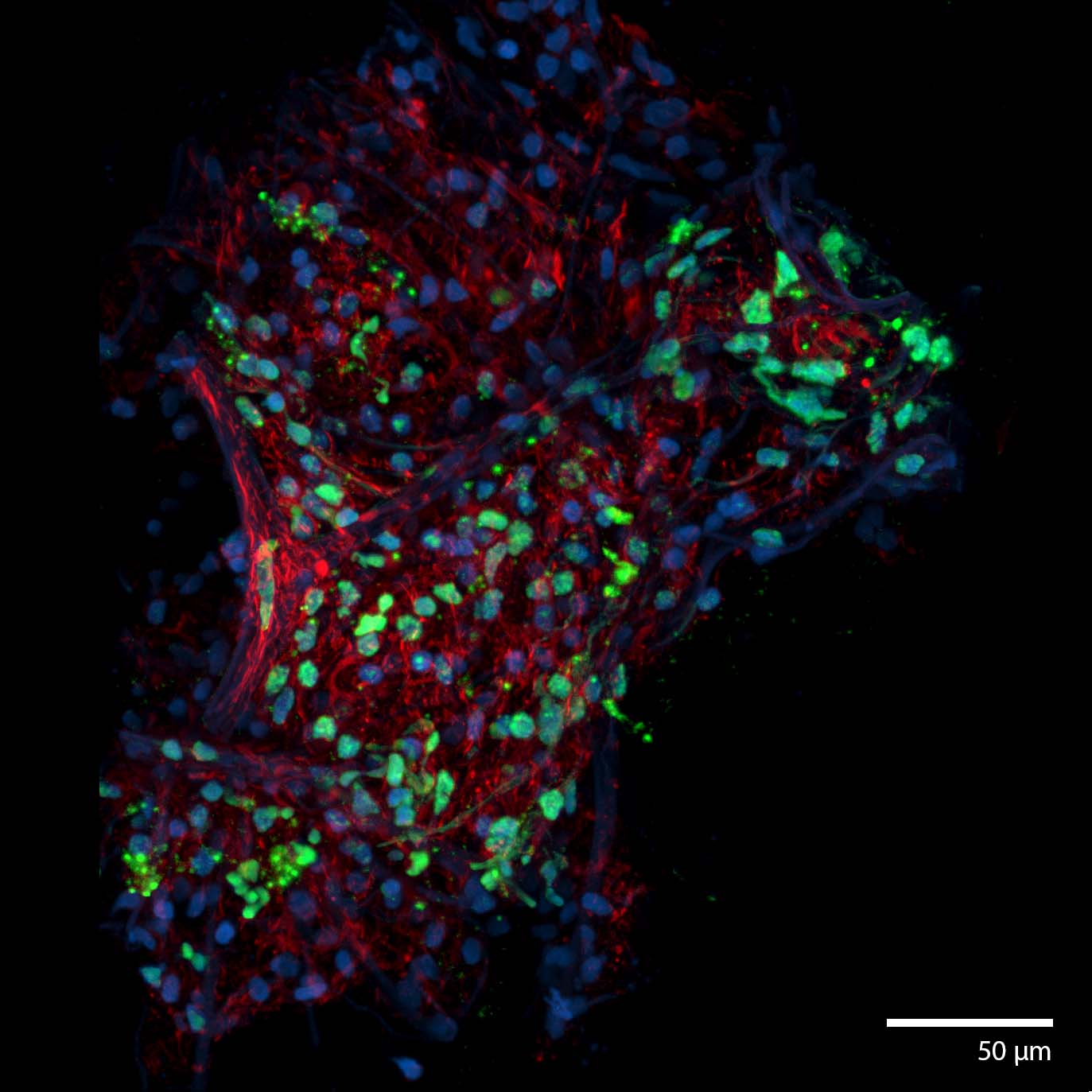
3D Printed osteosarcoma tumoroidThis patient derived osteosarcoma tumoroid was 3d printed and matured under perfusion in the 3D Darcy Plates with a background oxygen partial pressure of ~20%. At the time of imaging this sample was 14 days old. This sample was used during our studies in osteosarcoma and pediatric cancer. Mitotic figures and nuclear atypia can be seen through this tumoroid. The tumoroids maintained oncogenically active cells as can be seen through an oncogenic reported (FITC:green). This osteosarcoma tumoroid was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 14 days and stained for actin (TRITC: red) and the nuclei (DAPI: blue). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
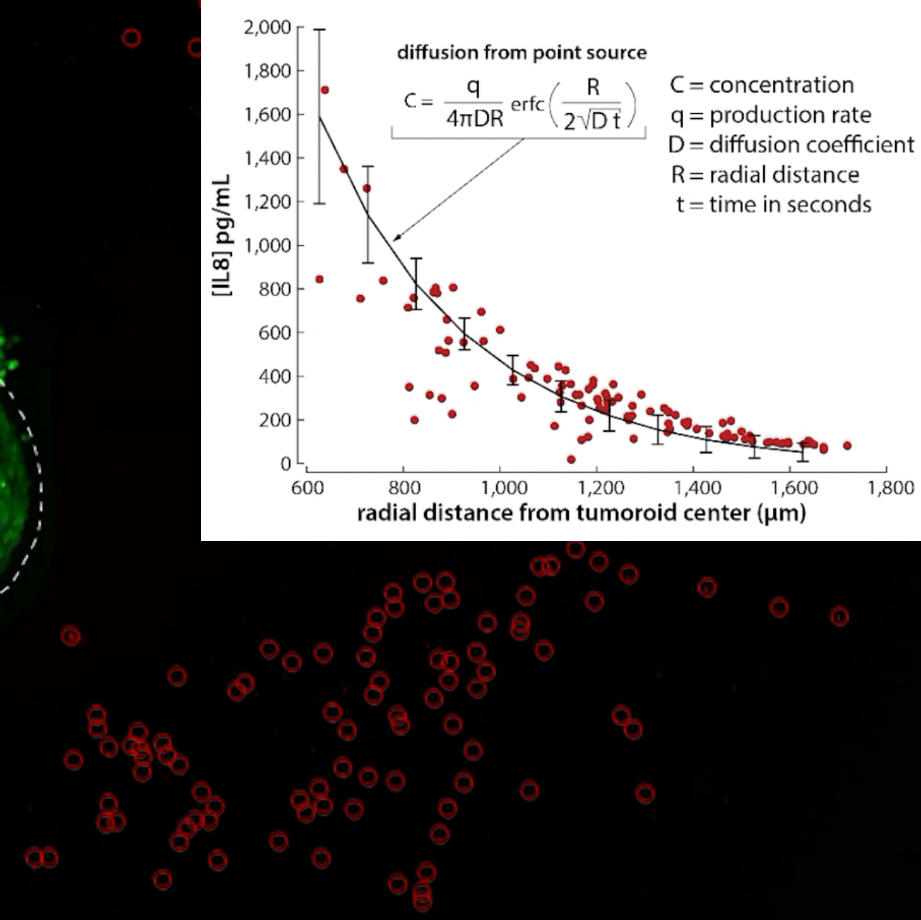 |
IN situ measurements of cytokine productionThe Liquid Like Solid (LLS) 3D Culture medium facilitates the positioning of solid objects, cells, and tissues. An osteosarcoma tumoroid (similar to above) is placed in the LLS with a field of ELISA beads that can be used to capture secreted cytokines. Osteosarcoma is well known to produce Interleukin 8 (IL8), which is a chemokine. Through a calibration procedure the fluorescence of the beads can be converted into a local concentration of IL8. The movie shows the spatiotemporal measurements of IL8 around the osteosarcoma tumoroid. These data can be fit to known solutions based on diffusion to solve for the surface concentration of IL8 at the tumor margin, as well as production rates of IL8 by the tumor. These experiments are not performed under perfusion; the LLS suppresses convention, and in the absence of perfusion demonstrates nearly ideal Fickian diffusion behavior. Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
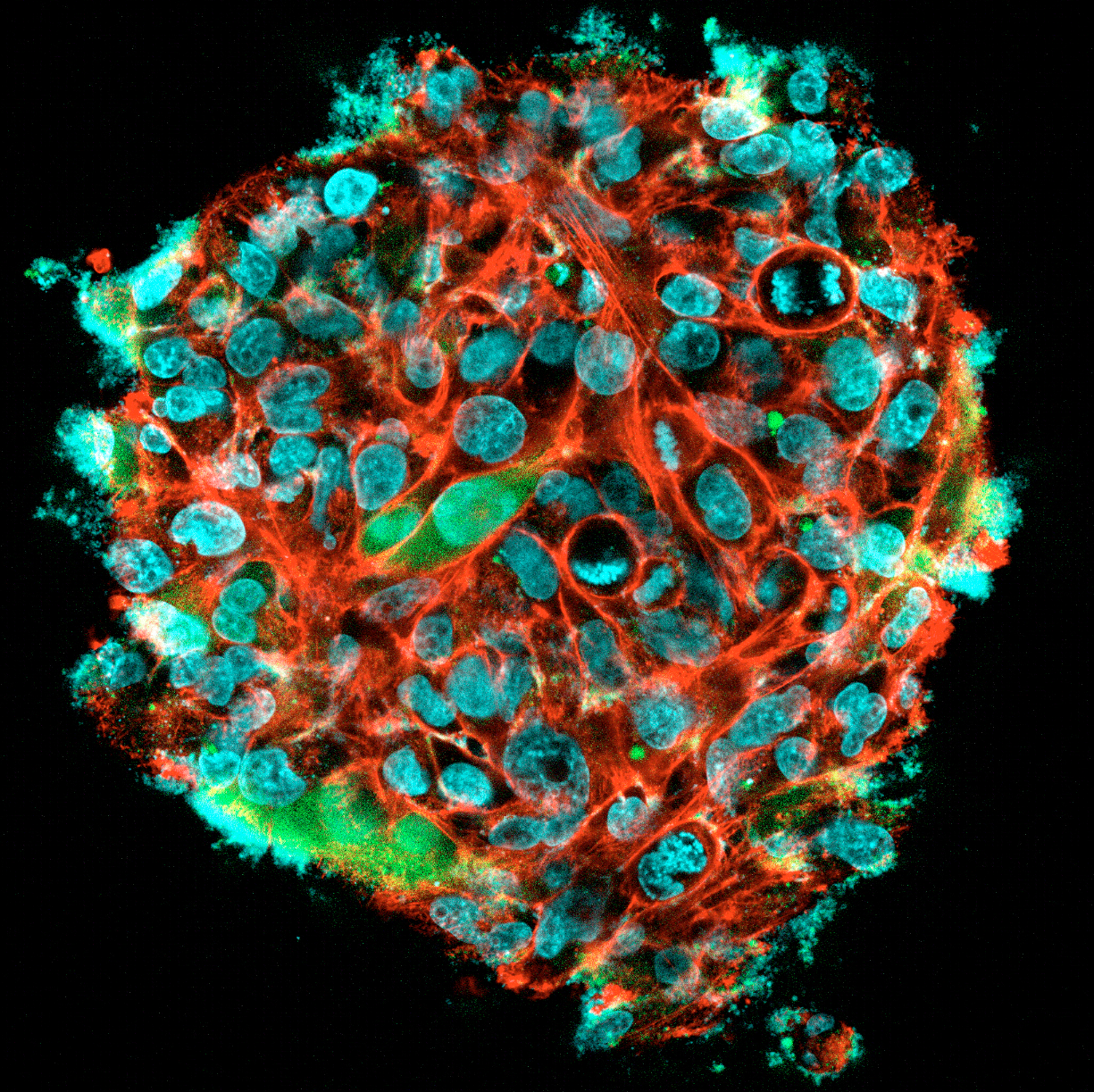
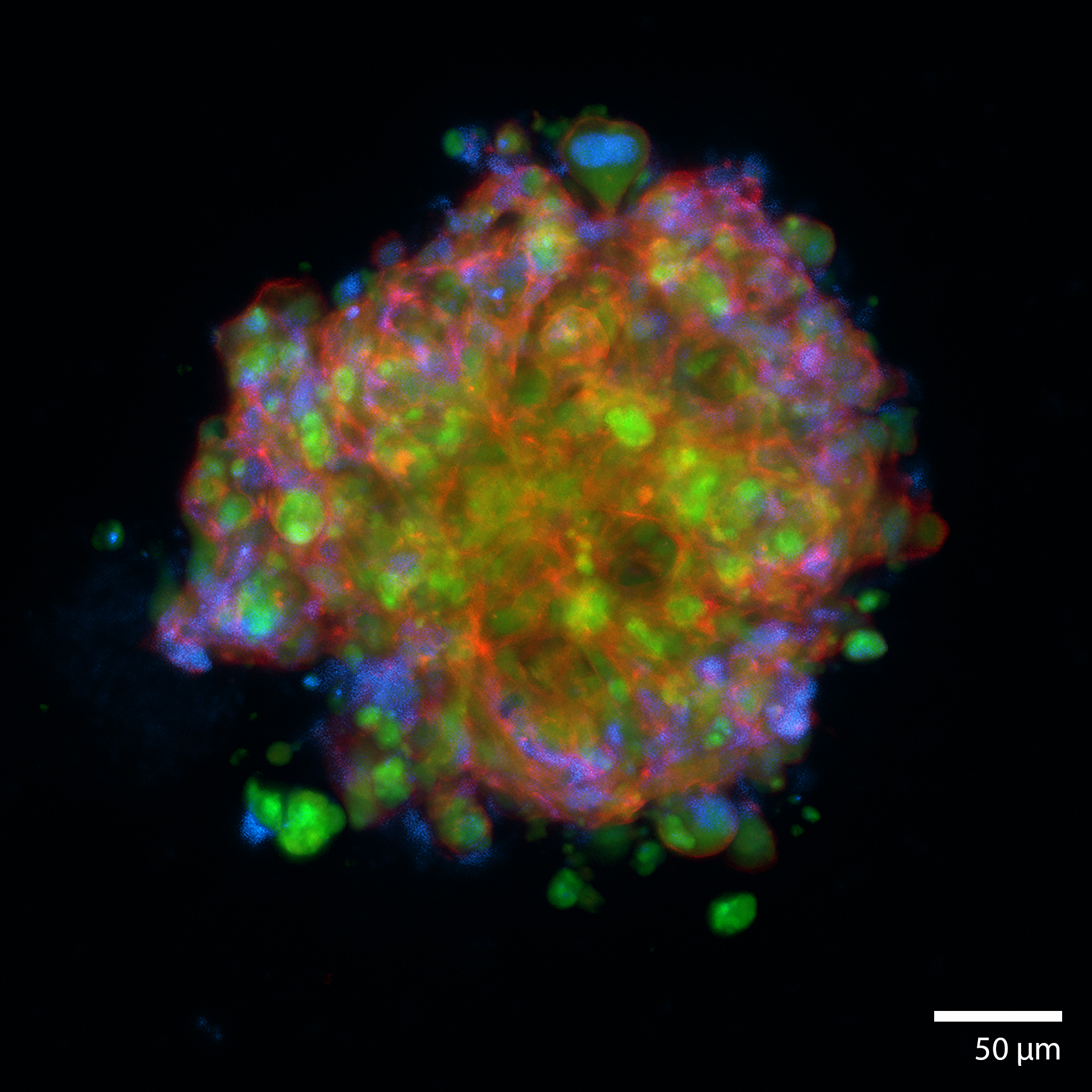
3D Pancreatic islet culture
|
 |
 |
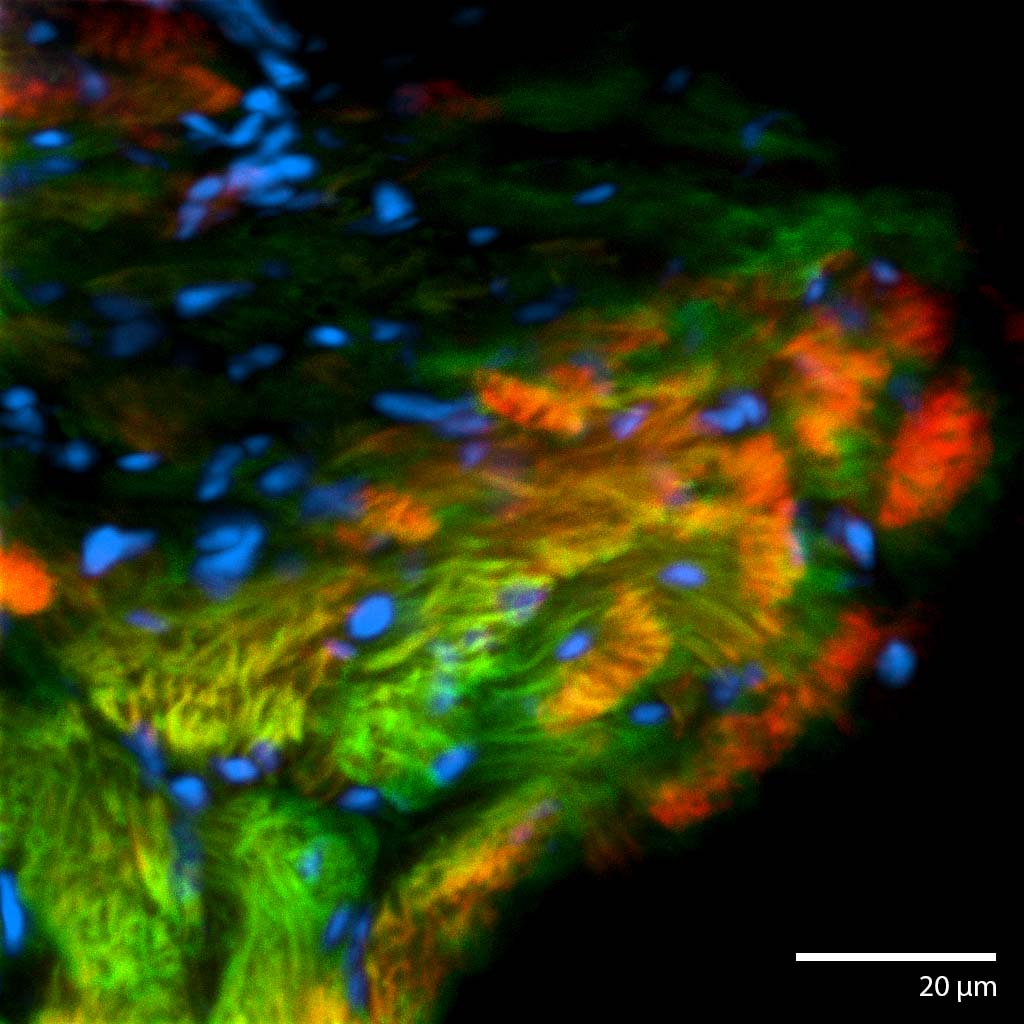
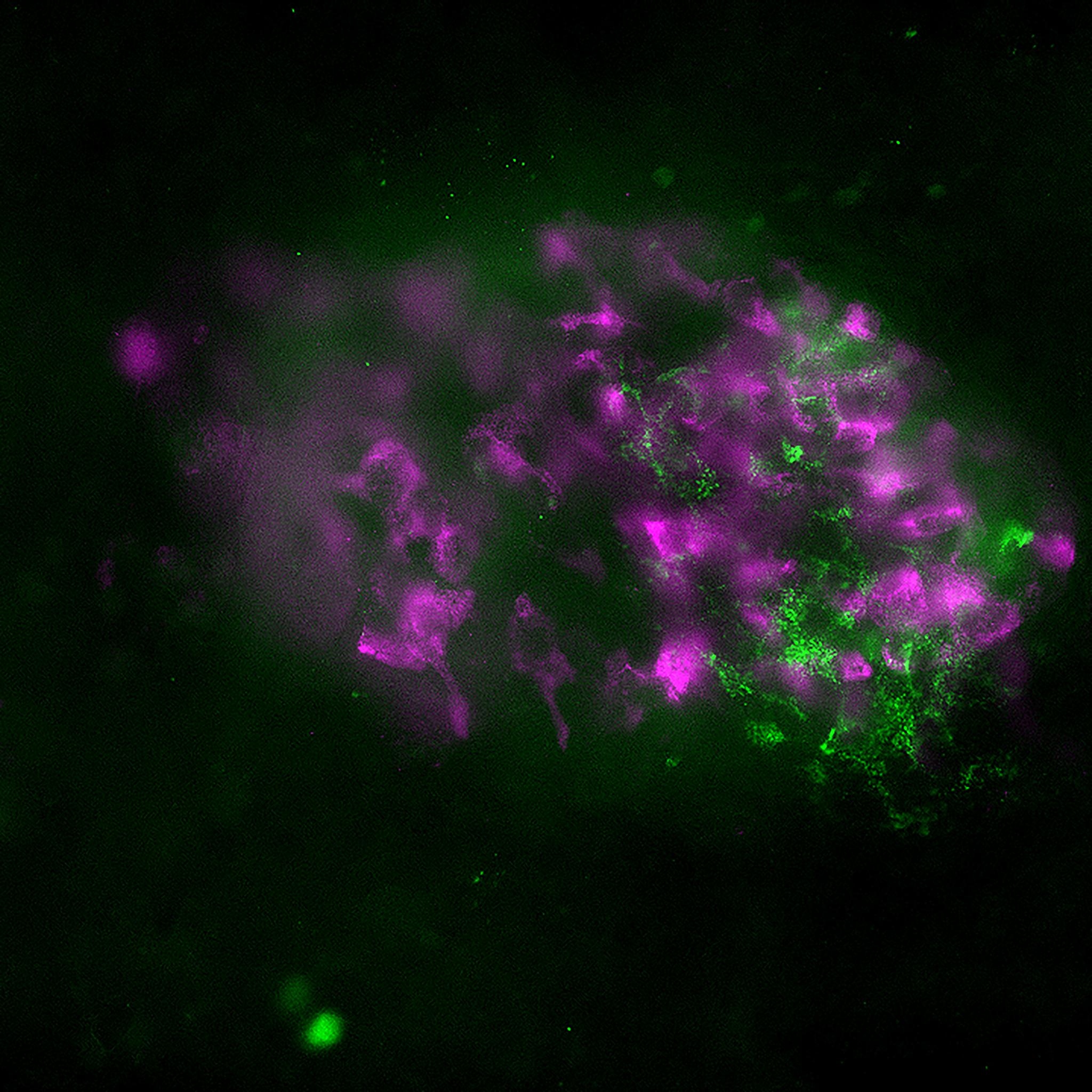
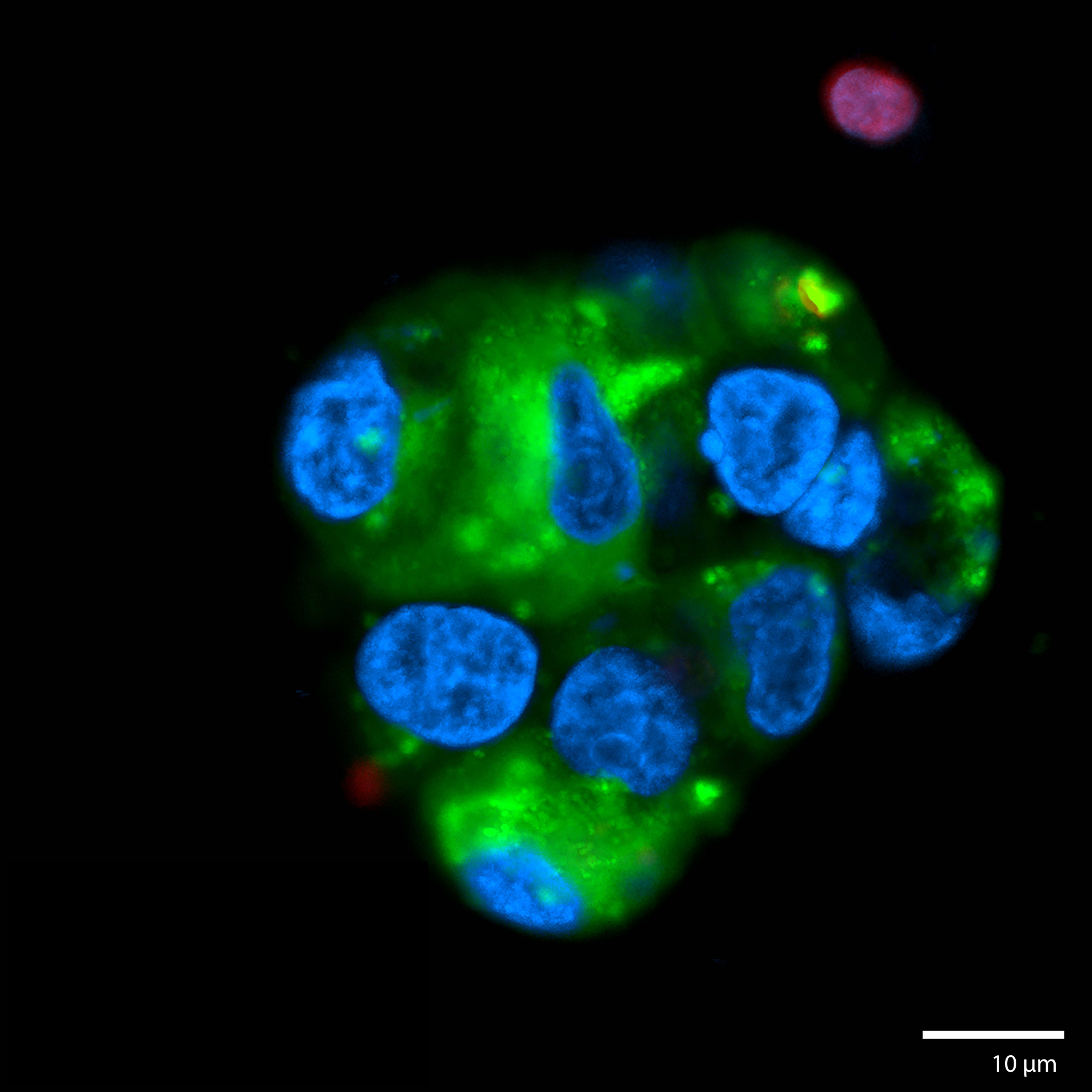
3D Printed Renal Cell Carcinoma tumoroidThis renal cell carcinoma tumoroid was 3d printed in Liquid Like Solid (LLS) 3D medium and matured under perfusion in the 3D Darcy Plates with a background oxygen partial pressure of ~20%. At the time of imaging this sample was 14 days old. This sample was used during our studies in immuno oncology and the role of cancer associated fibroblasts. These renal cell carcinoma tumoroids produce abundant extracellular matrix, which can be imaged and appears as a diffuse halo or corona around the tumoroids. Cancer associated fibroblasts (CAFs) readily attached to this ECM and rapidly coated and overgrew the tumoroids. This renal cell carcinoma tumoroid was fixed in place in the Liquid Like Solid (LLS) 3D culture medium after 14 days and stained for actin (TRITC:red), cell tracker CMFDA (FITC:green), and the nuclei were stained with the nuclei Hoechst 33342 (DAPI: blue). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
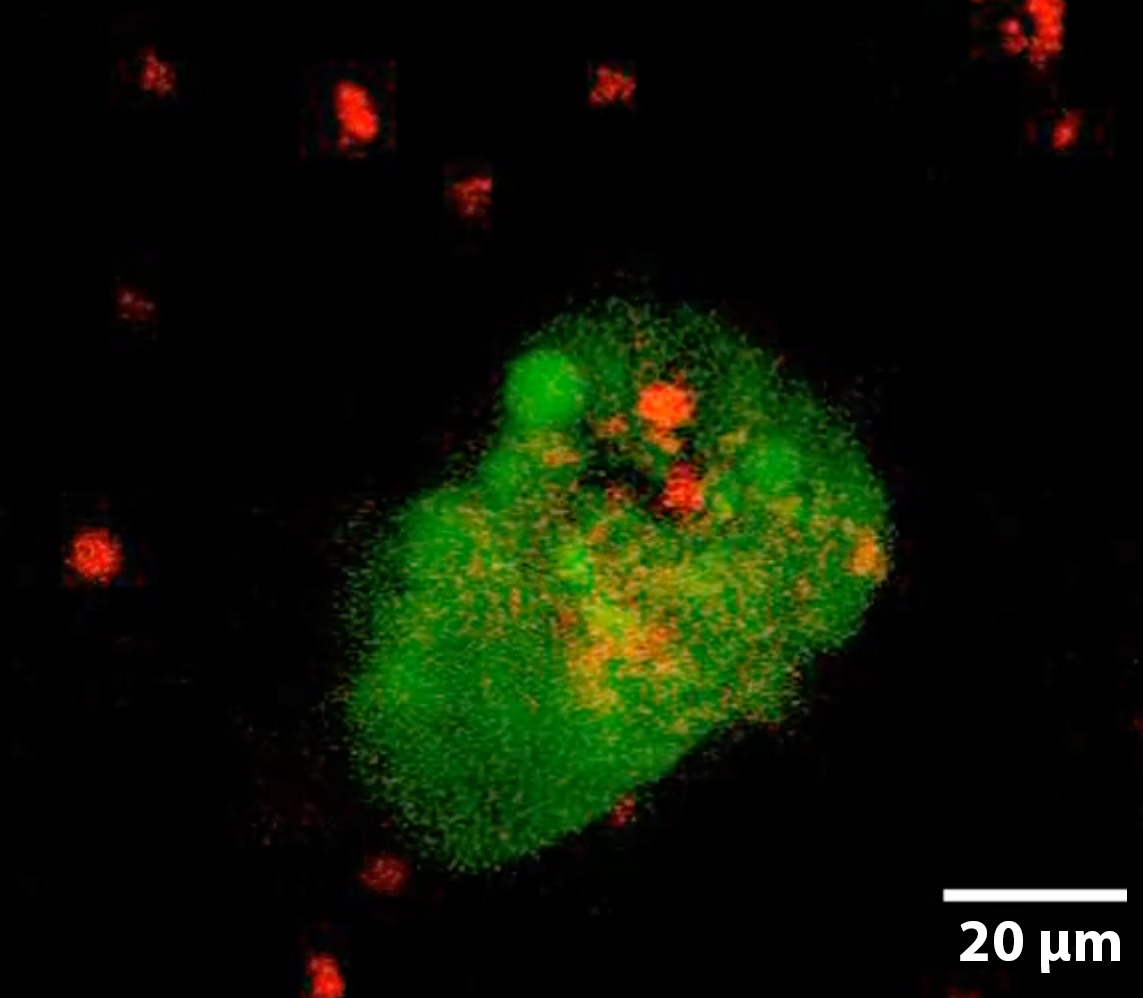
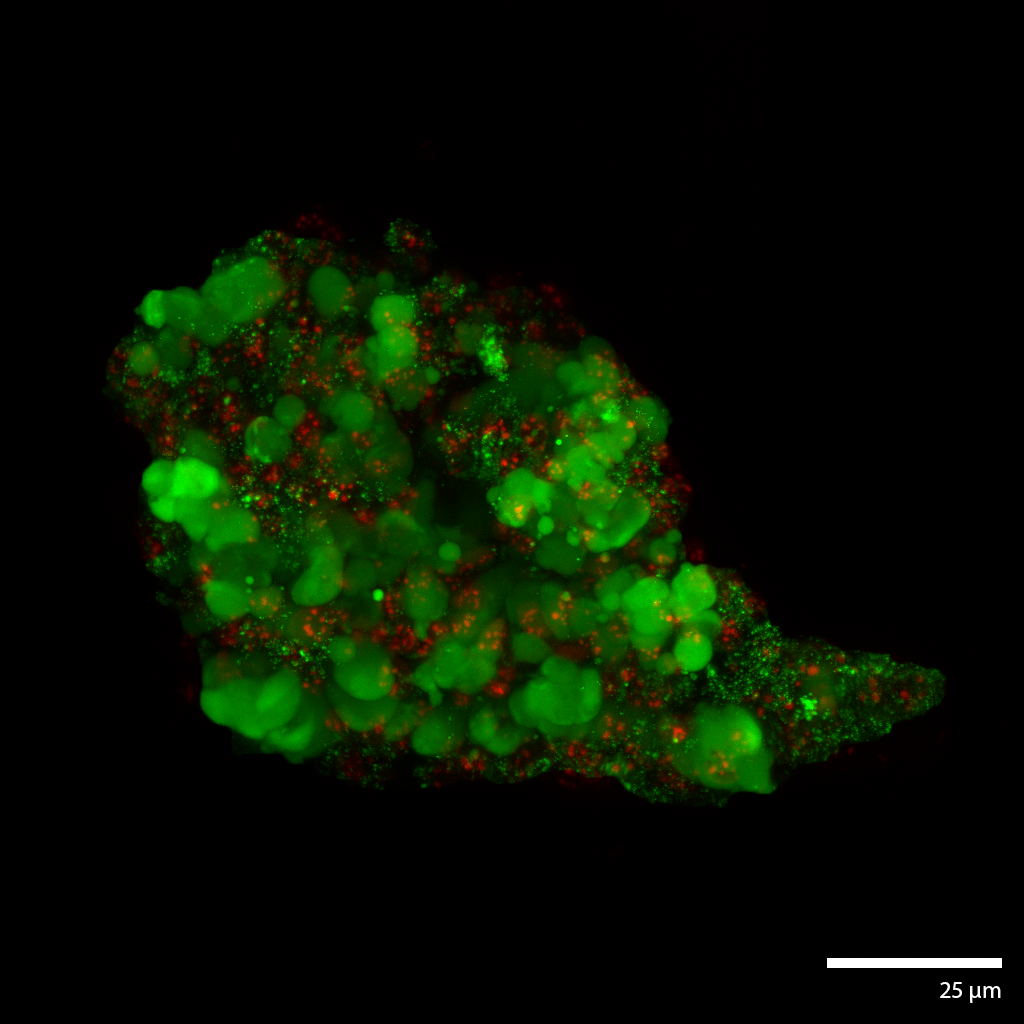
Hepatic Cancer micro-tumoroidsThis liver cancer micro-tumoroid develops from single cells dispersed throughout the interstitial space of the Liquid Like Solid (LLS) 3D culture medium. The incredibly large number of interstitial sites allow for 1×106 (million) cell culture in a single well of approximately 200 ul of LLS. Continuous perfusion in our Darcy plates, facilities continued growth and maturation over extended duration (weeks to months). Isolating the cells from LLS can be done through a number of techniques from elutriation to cell screening and density based liquid separation techniques. This hepatic cancer tumoroid was isolated and stained with a viability stain Calcein-AM (FITC: green), a nuclear stain Hoechst- 33342 (DAPI: blue), and a dead stain BOBO-3 Iodide (TRITC: red). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
 |
 |
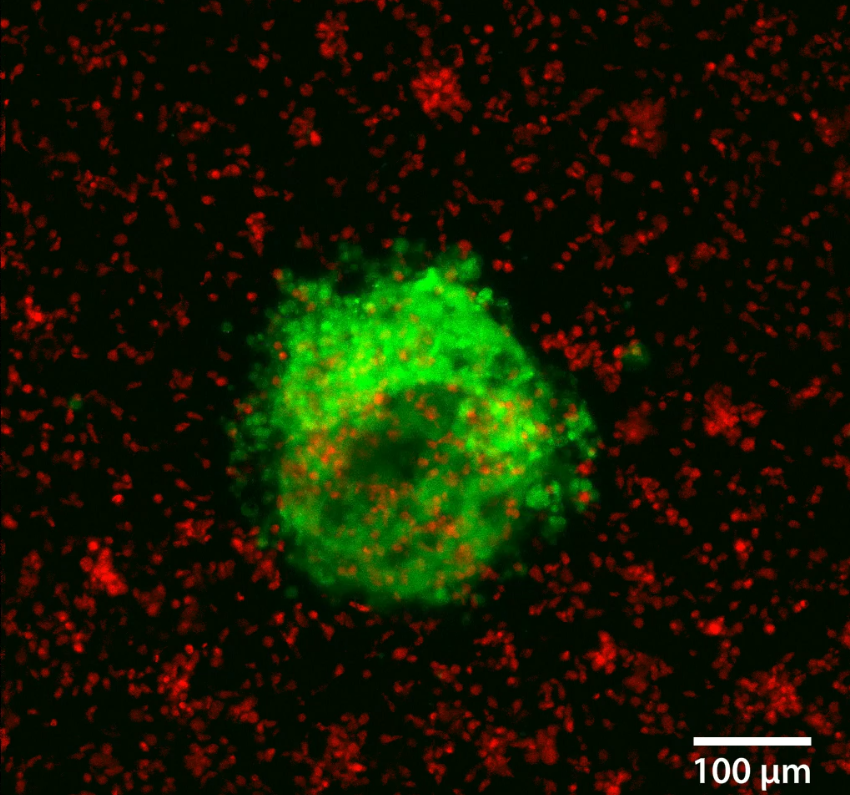
3D Human Colorectal Cancer TUmoroidThis human colorectal tumoroid was formed from a mechanically disaggregated sample taken from colorectal cancer surgical removal. The microtissue fragment was initially faceted as a result of the mechanical disaggregation and has retained some of this form during a maturation process under perfusion in the 3D Darcy Plates. At the time of imaging the sample was 14 days old, and had been under continuous perfusion in our Darcy plates at 90% background oxygen partial pressure. This sample was used during our immuno oncology in vitro experiments. Viability was confirmed out to 21 days, at which time the experiments were stopped; the samples were still functional and viable. These colorectal cancer tumoroid samples were roughly 100-400 um in diameter, and also produced copious amounts of mucin. This colorectal cancer tumoroid was isolated and stained with a viability stain Calcein-AM (FITC: green) and a dead stain BOBO-3 Iodide (TRITC: red). Image collected in the Cancer Engineering Laboratory on a fast scanning fluorescence confocal microscope, Nikon A1R. |
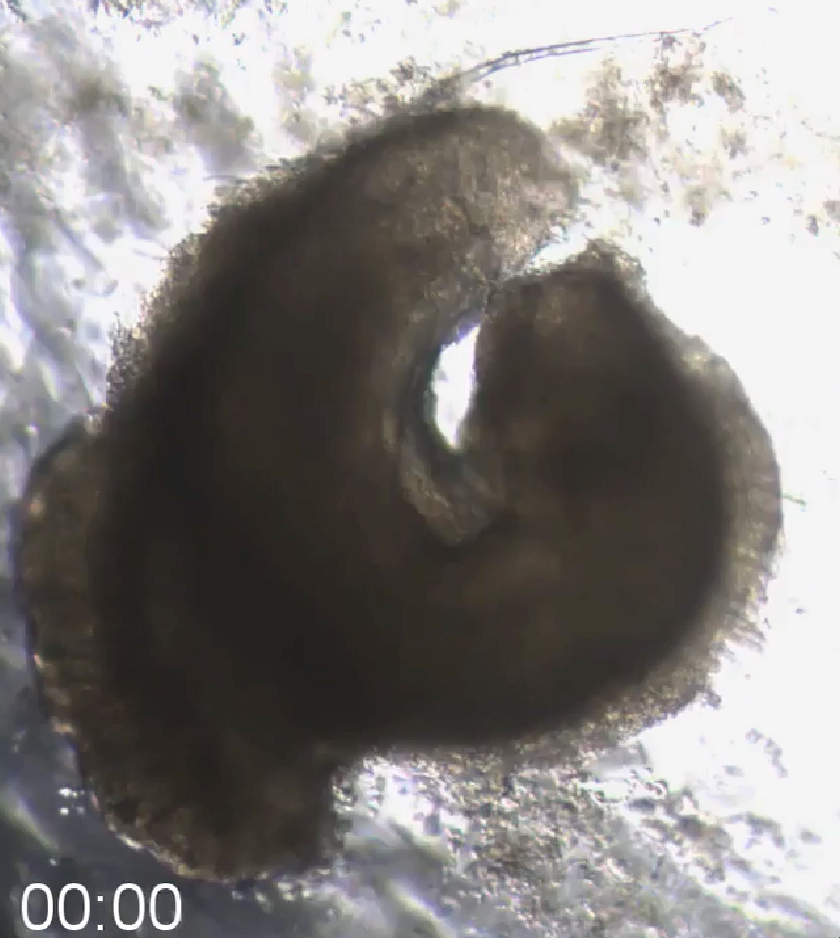
3D Colorectal microtissue explants and 3D OrganoidsThis colorectal organoid was formed from a mechanically disaggregated sample of the large intestine. The microtissue fragment retained its initially faceted morphology from mechanical disaggregation during a maturation process under perfusion in the 3D Darcy Plates. At the time of imaging the sample was 7 days old, and had been under continuous perfusion in our Darcy plates at 90% background oxygen partial pressure. This sample was used during an exploratory study of liquid culture medium tuning and optimization. These colorectal organoids and microtissue explants produced copious amounts of mucin, and some showed prolonged coordinated contractions over weeks in 3D culture. This image is from a brightfield microscopy imaging timelapse movie over 10 minutes. showing the continuous and persistent contractions, which responded to drugs, calcium channel blockers, and epinephrine. |
 |
 |
3D Printing in Liquid like solids
|